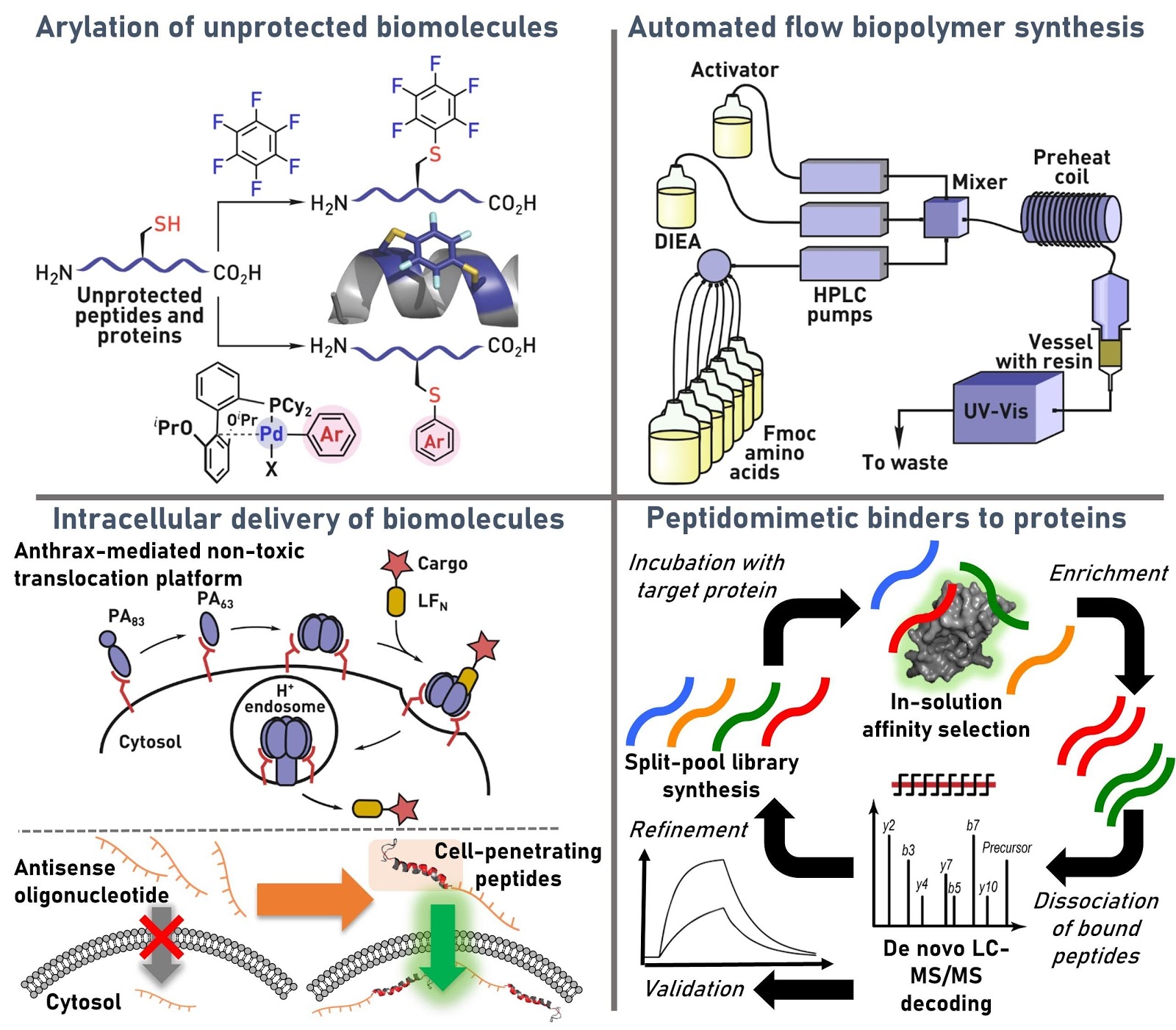
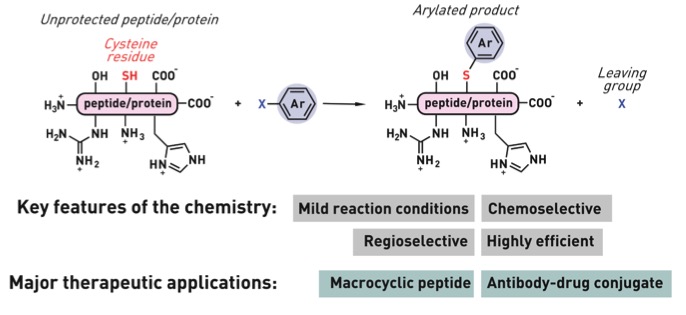
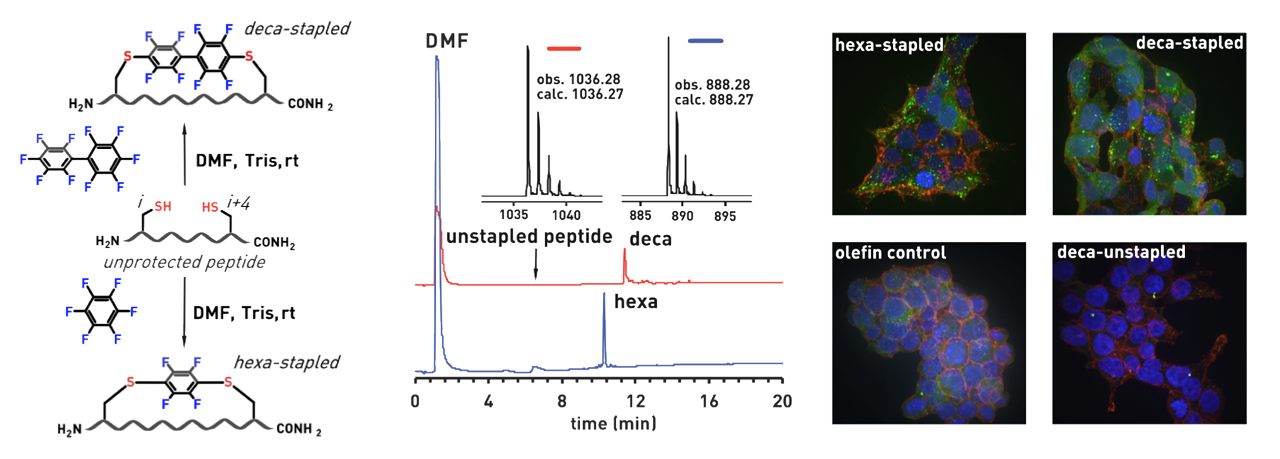
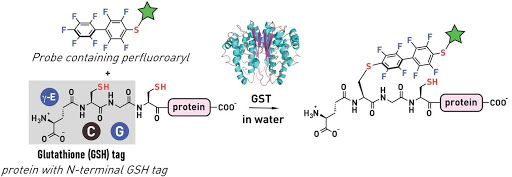
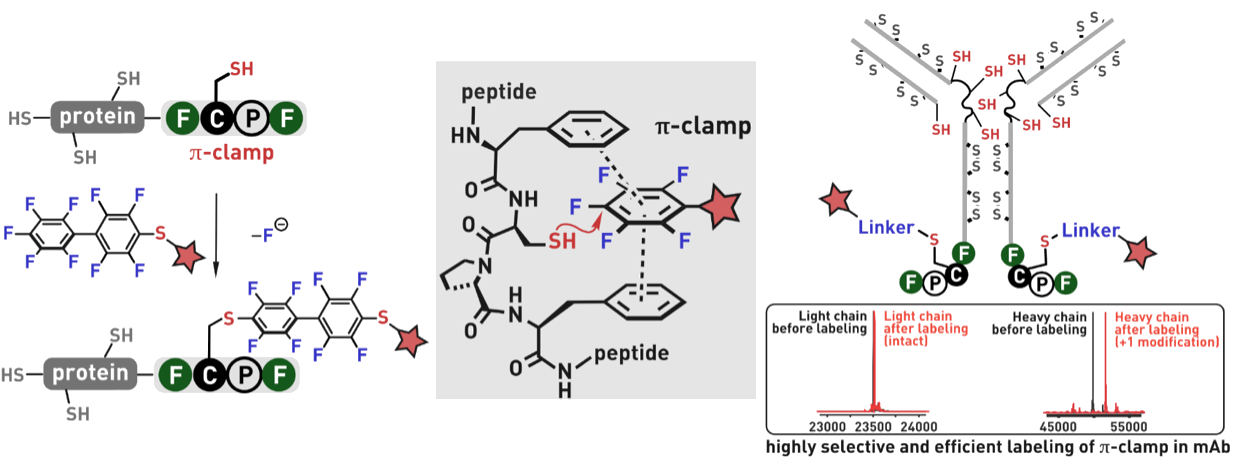
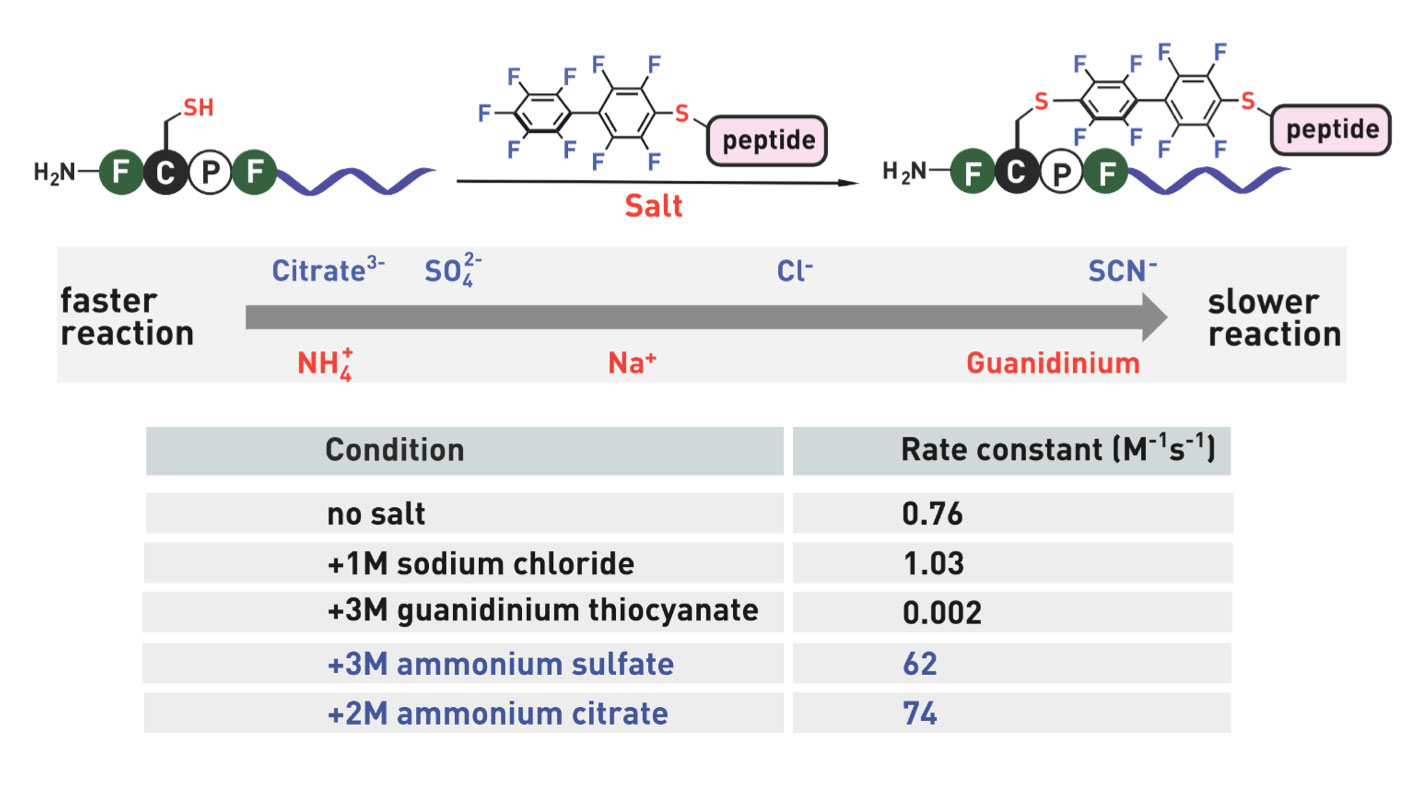
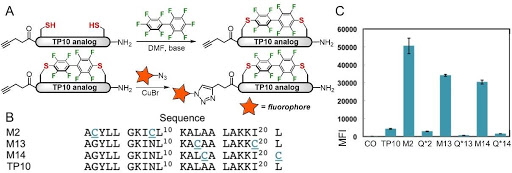
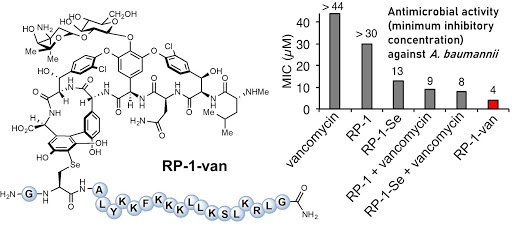
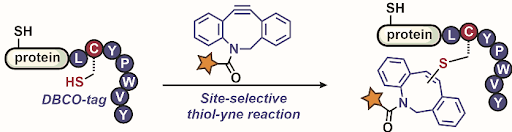
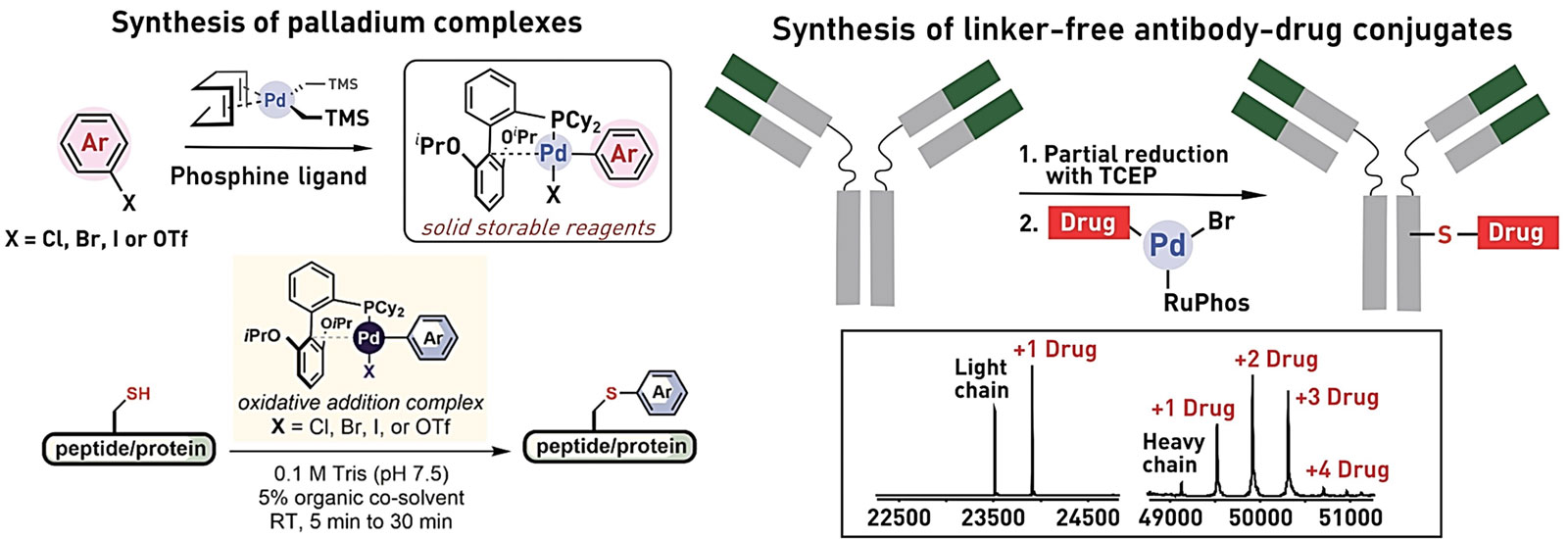
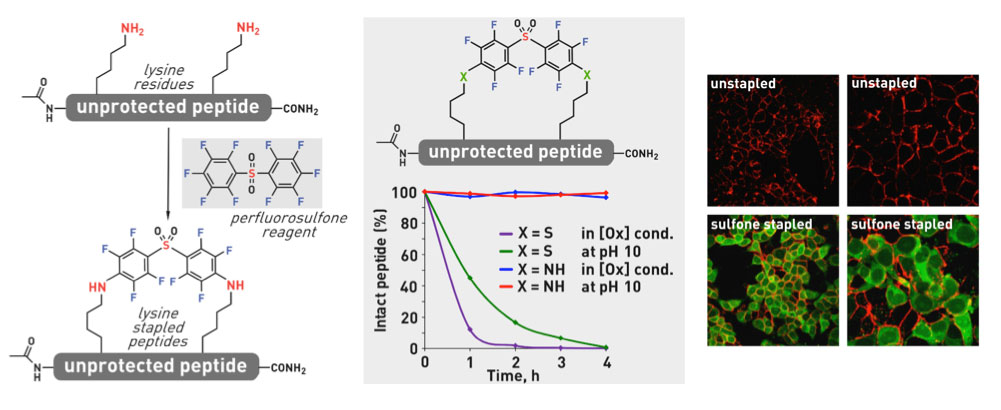
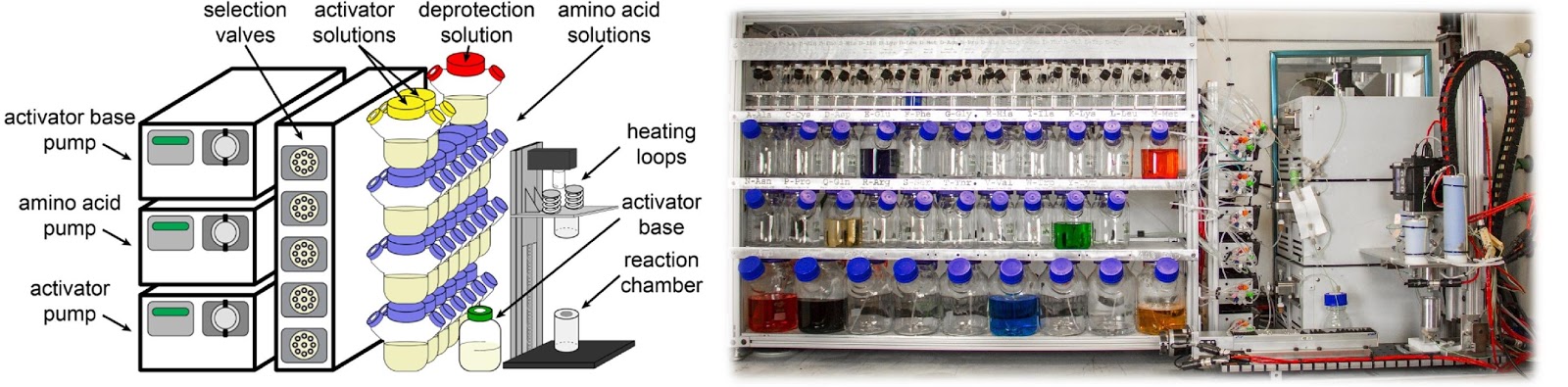
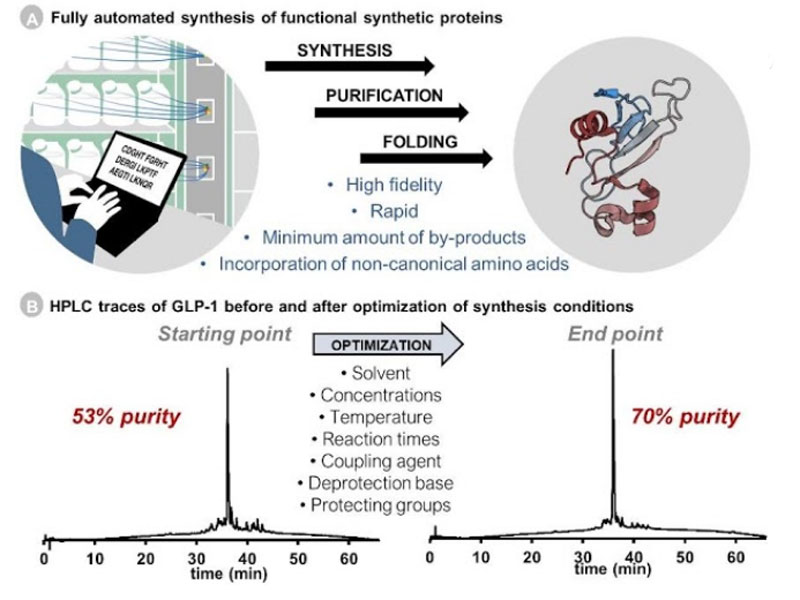
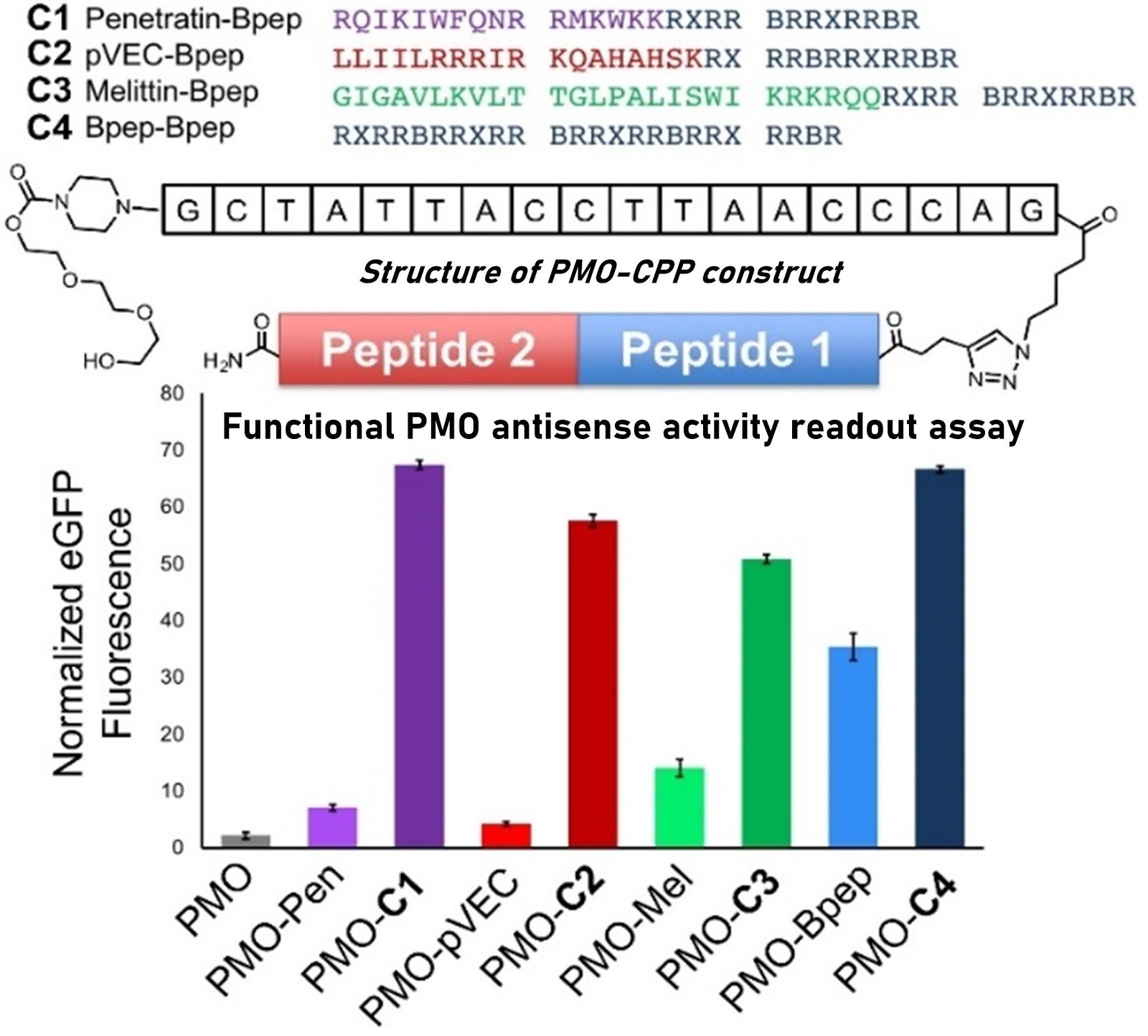
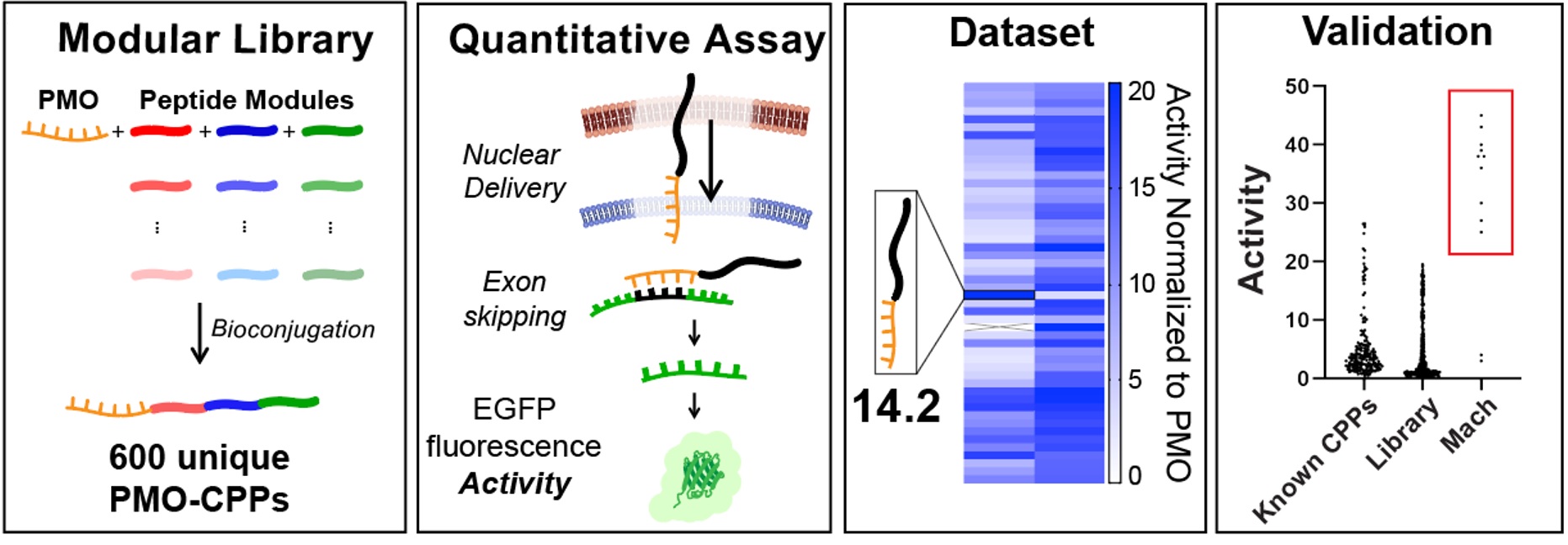
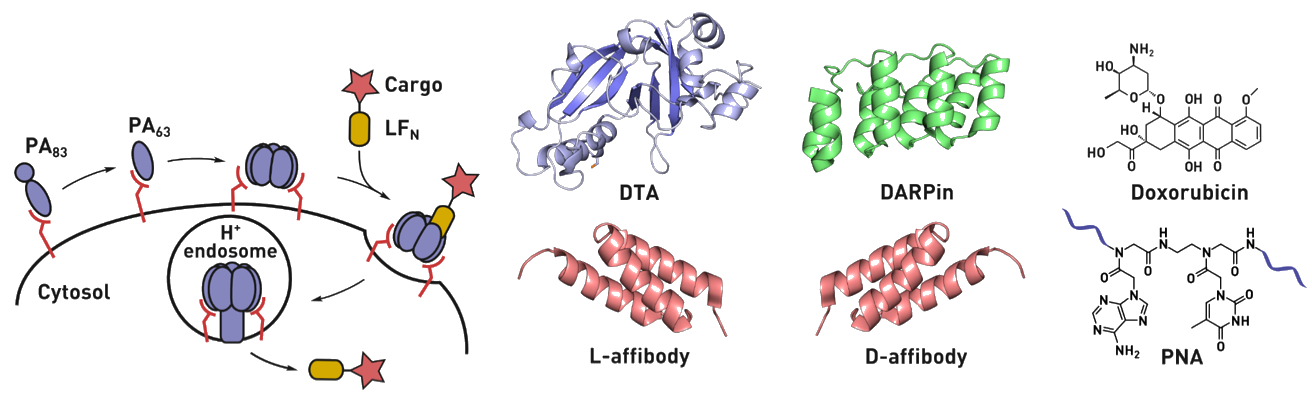
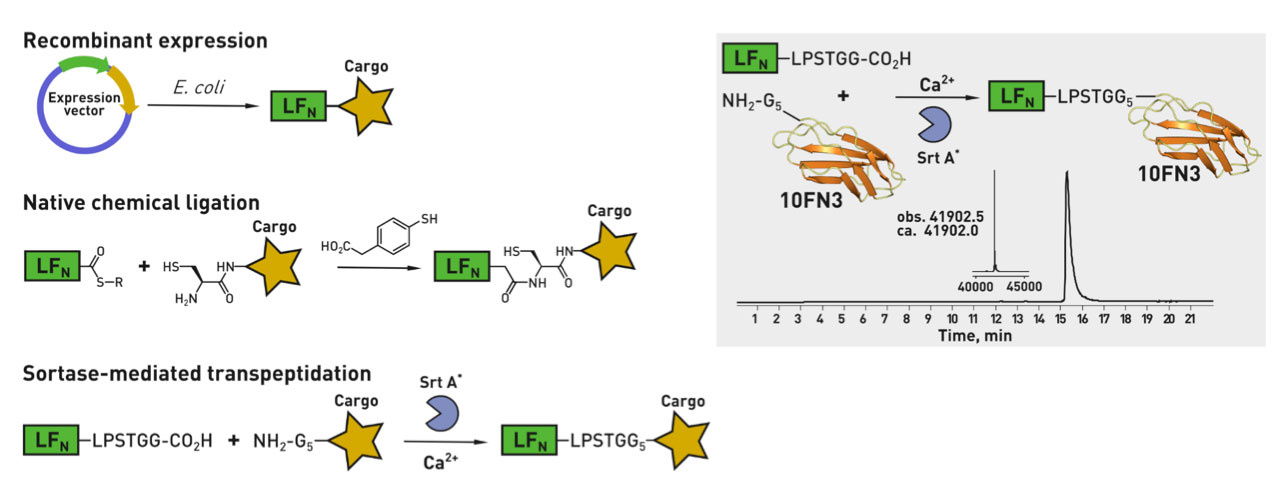
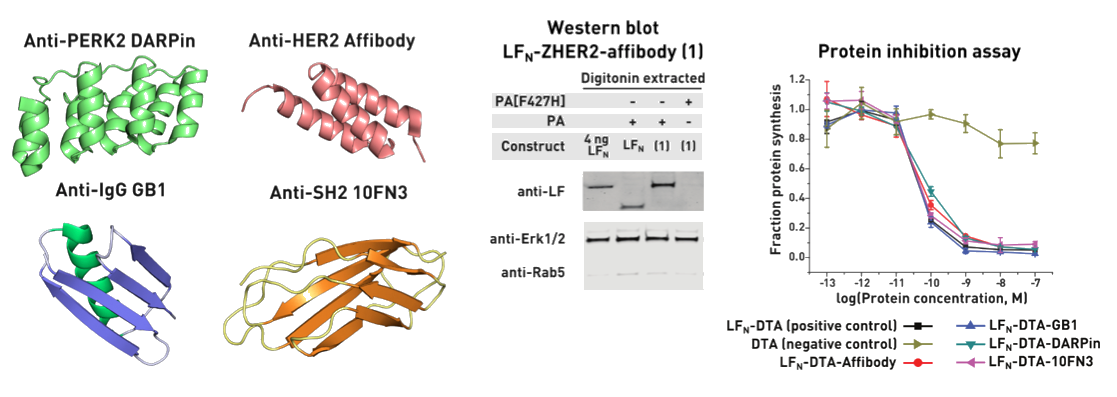
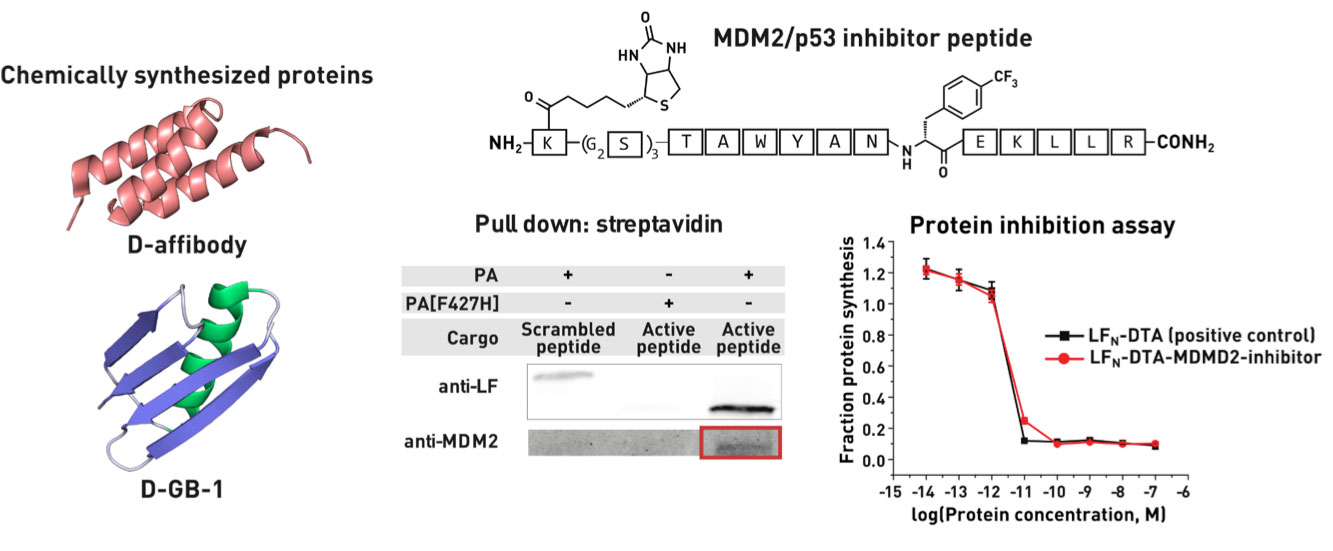
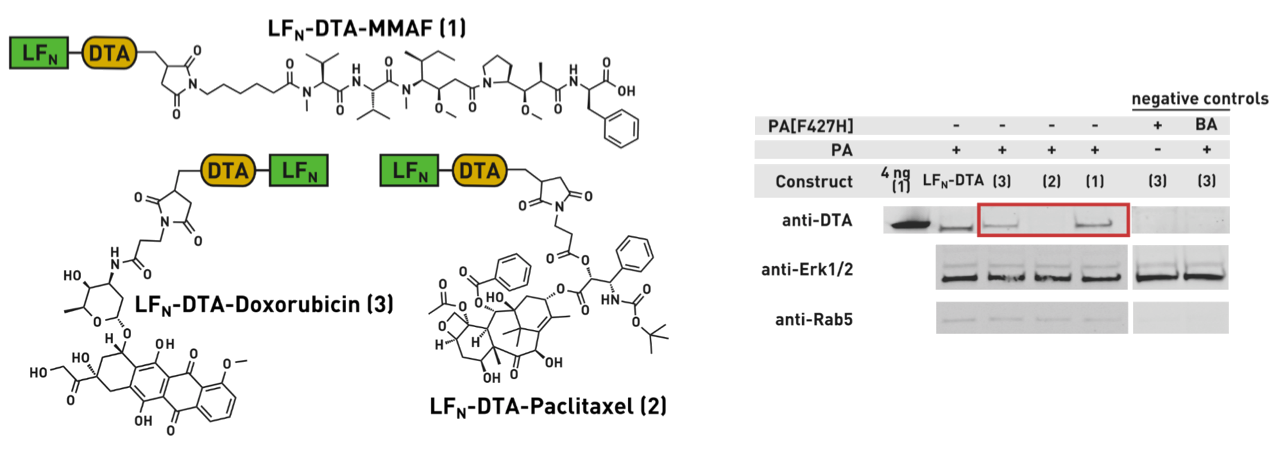
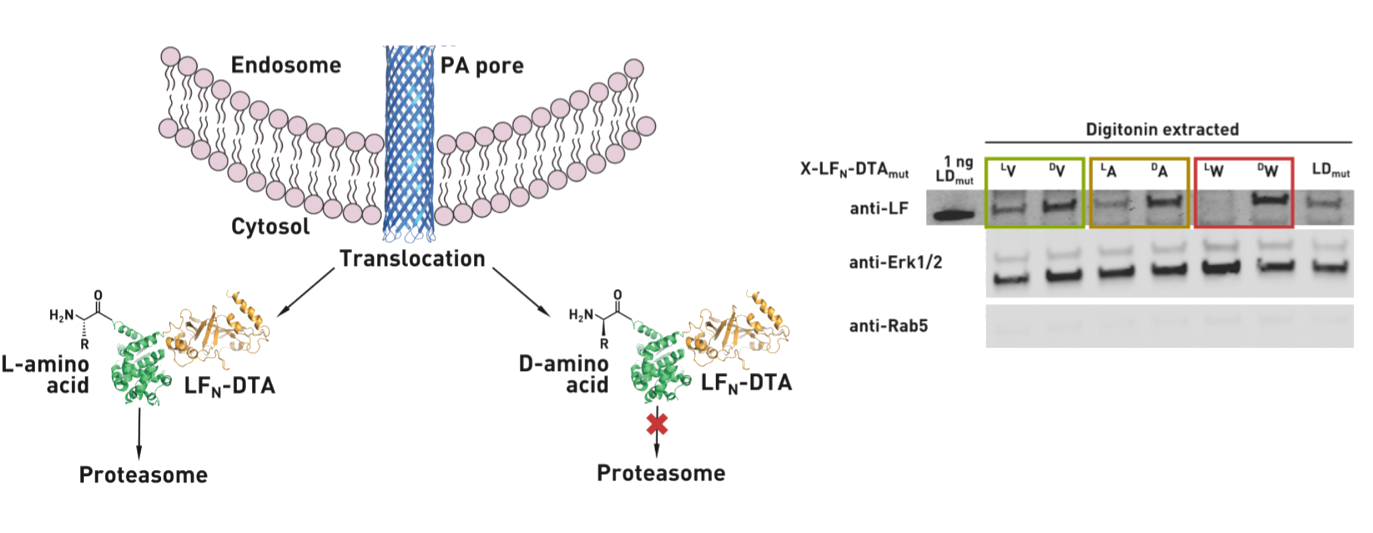
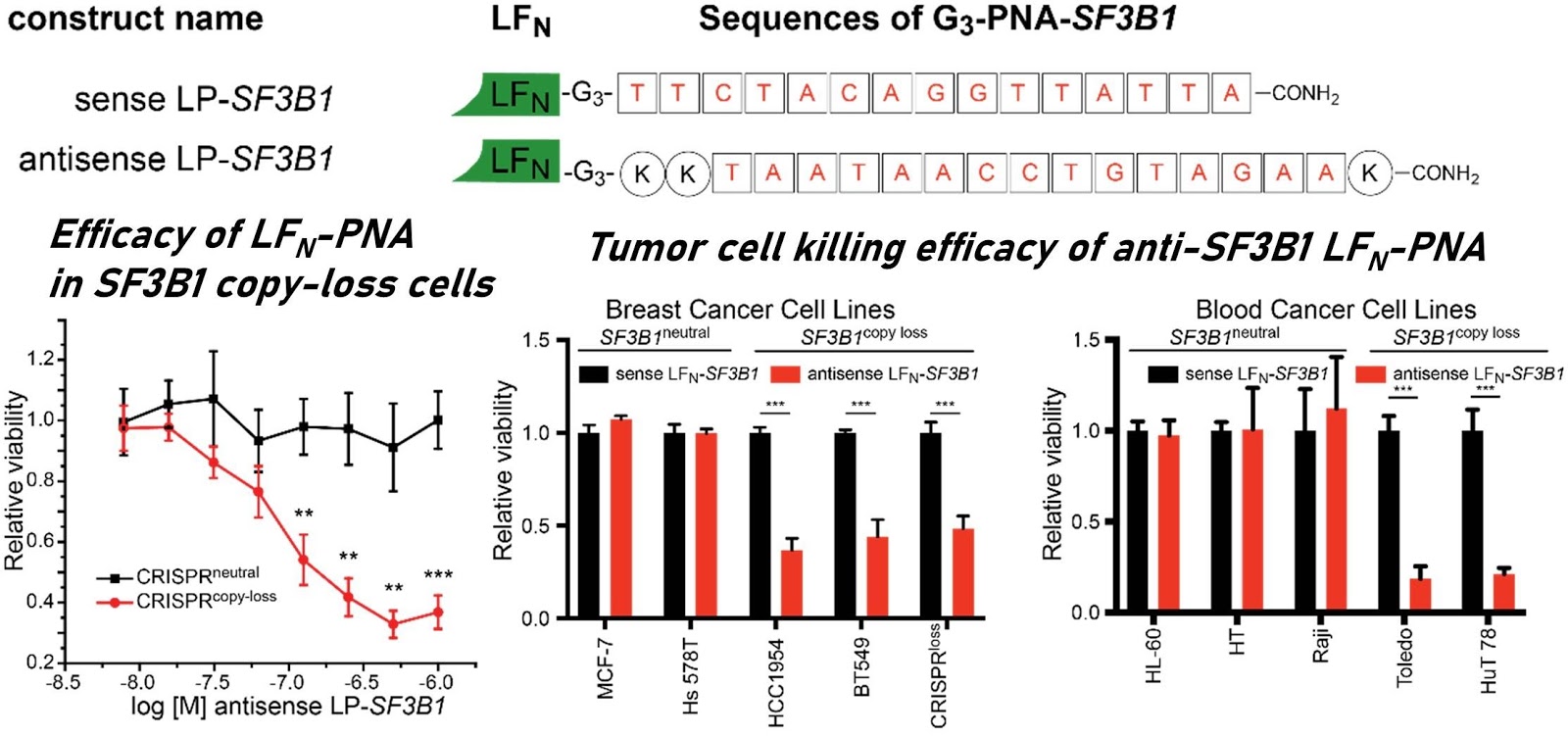
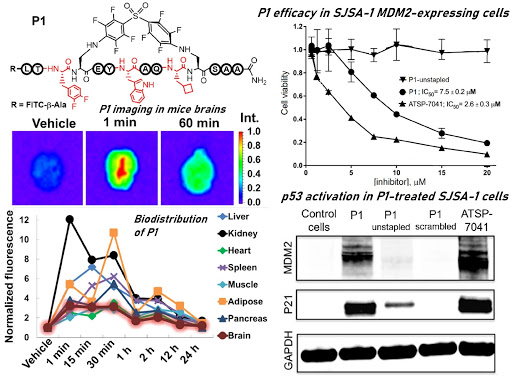
Ribosomes can produce proteins in minutes and are largely constrained to proteinogenic amino acids. We aim to develop high-fidelity multistep, continuous flow chemical processes for the efficient and rapid synthesis of biopolymers. These technologies provide an alternative for producing single-domain proteins without the ribosome and support our long-term goal of constructing a “chemical factory” in which starting reagents flow in and pure biomolecules of interest flow out. We invented a novel flow-based platform for the ultra-rapid production of synthetic peptides in collaboration with Klavs Jensen’s lab. This machine, termed automated fast-flow peptide synthesizer (AFPS), allows us to increase the rate of peptide production by a factor of 60 versus commercial peptide synthesizers, enabling amide coupling reactions in 10 seconds or less and performing a full amide coupling cycle in 1 minute. We used AFPS technology for expedited and high-fidelity production of personalized cancer vaccines for clinical partners and produced the majority of human antimicrobial peptides with AFPS instrumentation. Automated flow technology not only facilitates rapid biopolymer production, but has enabled us to carry out entire D-scans of small proteins to investigate their folding and functions. We recently used this technology to achieve stepwise total chemical synthesis of protein chains nearing 200 amino acids in length that were demonstrated to retain the structure and function of native variants obtained by recombinant expression. Our future efforts will be to adapt this technology to flow synthesis of antisense oligonucleotides, harness the power of machine learning to predict and eliminate aggregation, and combine flow synthesis of protein domains with native chemical ligation and protein folding, eventually arriving at an integrated system capable of synthesizing the majority of human proteins on demand. This technology has the potential to solve the manufacturing problem for custom or on-demand proteins and life-saving personalized therapies, and is amenable to designer chemistry with the use of a variety of backbones and building blocks, including D-amino acids, glycosylated functionalities, and non-canonical residues.
(M.D. Simon et al. ChemBioChem, 2014, 15, 713-20; R.L. Policarpo et al. Angew. Chem. Int. Ed., 2014, 53, 9203-8; S.K. Mong et al. ChemBioChem, 2014, 15, 721-33; T. Luhmann et al. Org. Biomol. Chem., 2016, 14, 3345-9; M.D. Simon et al. J. Am. Chem. Soc., 2016, 138, 12099-111; A.J. Mijalis et al. Nat. Chem. Biol., 2017, 13, 464-6; N.L. Truex et al. Sci. Rep., 2020, 10, 723; J.S. Albin and B.L. Pentelute, Aust. J. Chem., 2020, 73, 380-388; N. Hartrampf et al. Science, 2020, 368, 980-987)