
Design, Structure–Activity Relationships, and Computational Modeling Studies of a Series of α-Helix Biased, Ultra-Short Glucagon-like Peptide-1 Receptor Agonists
Design, Structure–Activity Relationships, and Computational Modeling Studies of a Series of α-Helix Biased, Ultra-Short Glucagon-like Peptide-1 Receptor Agonists
Jonathon R. Sawyer, Joseph A. Audie, Jon Swanson, David Diller, Solimar Santiago, Valentin K. Gribkoff, Allison Ackerman, Victor J. Hruby, Gianpaolo Gobbo, Michael A. Bellucci, William A. Glauser, Brad L. Pentelute andTomi K. Sawyer
Abstract
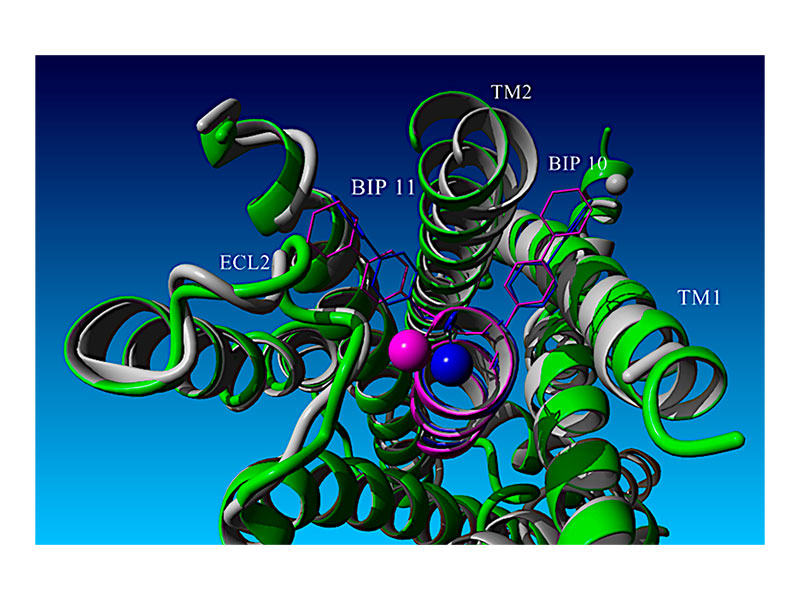
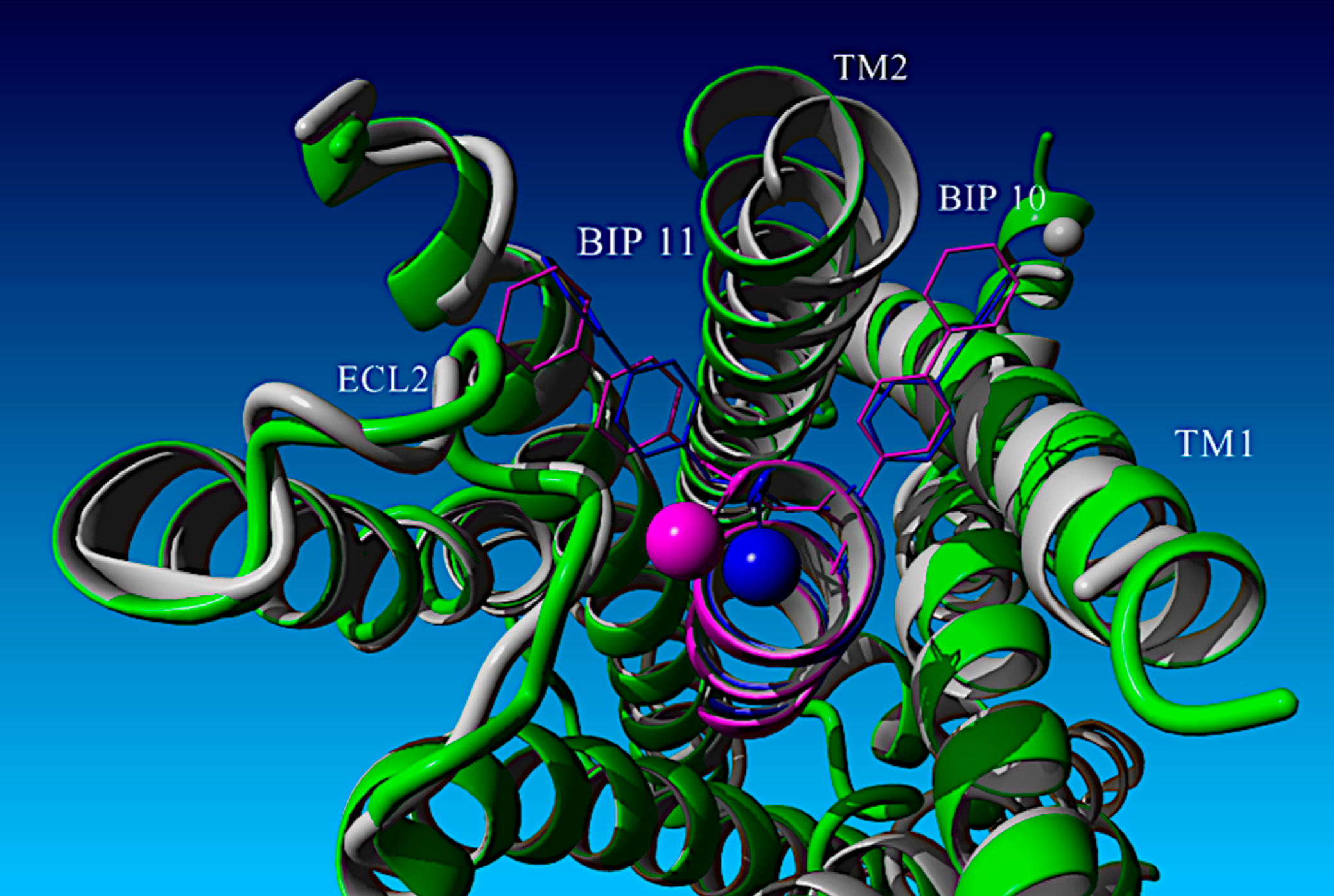
A systematic structure–activity and computational modeling analysis of a series of glucagon-like peptide-1 receptor (GLP-1R) agonists based upon an ultra-short GLP-1 peptide, H-His-Aib-Glu-Gly-Thr-Phe-Thr-Ser-Asp-Bip-Bip-NH2, was conducted. This highly potent 11-mer peptide led to a deeper understanding of the α-helical bias of strategic α-methylation within the linear parent template as well as optimization of GLP-1R agonist potency by 1000-fold. These data were correlated with previously reported co-structures of both full-length GLP-1 analogs and progenitor N-terminal GLP-1 fragment analogs related to such ultra-short GLP-1R agonist peptides. Furthermore, the development of a quantitative structure–activity relationship (QSAR) model to analyze these findings is described in this study.



