
Deep Learning for Prediction and Optimization of Fast-Flow Peptide Synthesis
Deep Learning for Prediction and Optimization of Fast-Flow Peptide Synthesis
ACS Cent. Sci. 2020, 6, 12, 2277–2286
Publication Date:November 12, 2020
https://doi.org/10.1021/acscentsci.0c00979
Somesh Mohapatra, Nina Hartrampf, Mackenzie Poskus, Andrei Loas, Rafael Gómez-Bombarelli*, and Bradley L. Pentelute
Abstract
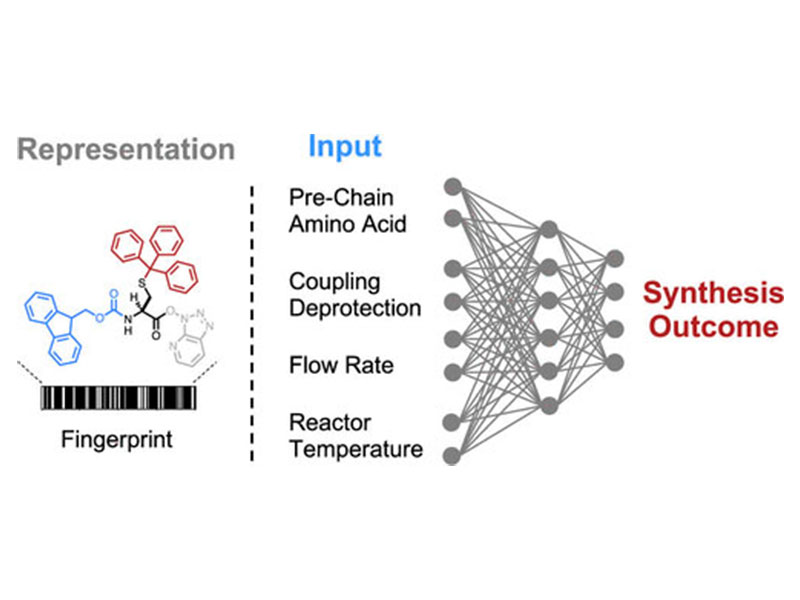
The chemical synthesis of polypeptides involves stepwise formation of amide bonds on an immobilized solid support. The high yields required for efficient incorporation of each individual amino acid in the growing chain are often impacted by sequence-dependent events such as aggregation. Here, we apply deep learning over ultraviolet–visible (UV–vis) analytical data collected from 35 427 individual fluorenylmethyloxycarbonyl (Fmoc) deprotection reactions performed with an automated fast-flow peptide synthesizer. The integral, height, and width of these time-resolved UV–vis deprotection traces indirectly allow for analysis of the iterative amide coupling cycles on resin. The computational model maps structural representations of amino acids and peptide sequences to experimental synthesis parameters and predicts the outcome of deprotection reactions with less than 6% error. Our deep-learning approach enables experimentally aware computational design for prediction of Fmoc deprotection efficiency and minimization of aggregation events, building the foundation for real-time optimization of peptide synthesis in flow.



