
Assessing the Performance of Peptide Force Fields for Modeling the Solution Structural Ensembles of Cyclic Peptides
Assessing the Performance of Peptide Force Fields for Modeling the Solution Structural Ensembles of Cyclic Peptides
Jiayuan MiaoArghya Pratim GhoshMinh Ngoc HoChengxi LiXuejian HuangBradley L. PenteluteJames D. BalejaYu-Shan Lin*
Abstract
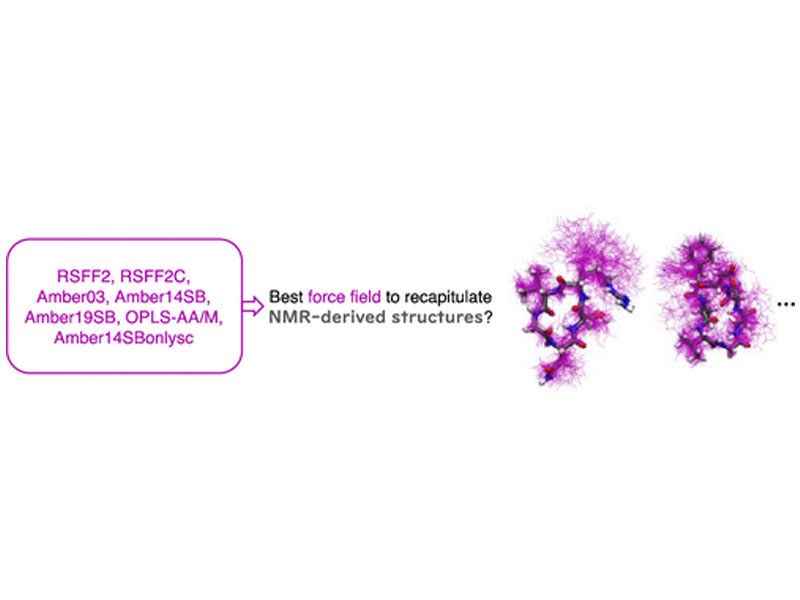
Molecular dynamics simulation is a powerful tool for characterizing the solution structural ensembles of cyclic peptides. However, the ability of simulation to recapitulate experimental results and make accurate predictions largely depends on the force fields used. In our work here, we evaluate the performance of seven state-of-the-art force fields in recapitulating the experimental NMR results in water of 12 benchmark cyclic peptides, consisting of 6 cyclic pentapeptides, 4 cyclic hexapeptides, and 2 cyclic heptapeptides. The results show that RSFF2+TIP3P, RSFF2C+TIP3P, and Amber14SB+TIP3P exhibit similar and the best performance, all recapitulating the NMR-derived structure information on 10 cyclic peptides. Amber19SB+OPC successfully recapitulates the NMR-derived structure information on 8 cyclic peptides. In contrast, OPLS-AA/M+TIP4P, Amber03+TIP3P, and Amber14SBonlysc+GB-neck2 could only recapitulate the NMR-derived structure information on 5 cyclic peptides, the majority of which are not well-structured.



