
A reactive peptide interface for site-selective cysteine bioconjugation
A reactive peptide interface for site-selective cysteine bioconjugation
https://doi.org/10.1039/D1CC00095K
Suan Tuang, Diomedes Dieppa-Matos, ORCID logo Chi Zhang, ORCID logo Christopher R. Shugrue, Peng Dai, Andrei Loas and Bradley L. Pentelute
Abstract
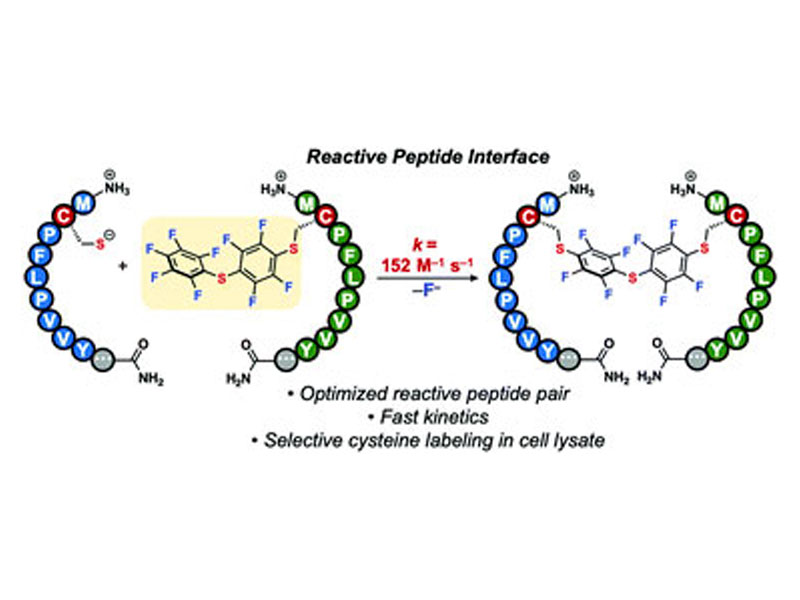
We report aqueous, site-selective modification of proteins using a reactive peptide interface comprising a nine-residue sequence. This interface is the fastest (second-order rate constant of 152 M−1 s−1) catalyst-free, cysteine-based method for modifying proteins available to date, and enables near-quantitative labeling of antibodies in cell lysate.
Graphical abstract: A reactive peptide interface for site-selective cysteine bioconjugation
Bioconjugation chemistry enables the development of new therapeutics,1,2 biomaterials,3,4 and probes for the exploration of complex biological processes.5 The site-selective modification of proteins, however, is challenging due to the chemical complexity of biomolecules and necessity for aqueous environments. Site-selective bioconjugation methods have been developed to address this challenge, some of which include unnatural amino acid incorporation into proteins,6–9 fusion of engineered, self-modifying enzymes,10,11 ligand-directed protein modification,12–14 and peptide tag-based modification.12–20
Peptide tags constitute a powerful approach to protein modification given that they are (1) small in size, making them less likely to perturb protein structure and function, and (2) usually composed of canonical amino acids, making them easier to genetically encode. The use of peptide sequences that accomplish otherwise sluggish reactions in water has been prevalent in the literature in this past decade.17–24 It is often hypothesized that the driving force for these reactions is the peptide's unique sequence. This sequence could afford improved reaction rates through enhanced nucleophilicity of the reactive residue arising from side-chain interactions and/or interactions between the peptide and the electrophile. Synthetic peptide libraries can be used to rapidly develop sequences that facilitate site-selective bionconjugations.21–24 Screening such libraries has led to the discovery of peptides that react preferentially with a specific electrophile.
Cysteine is commonly leveraged as the reactive residue in peptide tags due to its inherent nucleophilicity. Various Cys-containing peptides have been developed that react with electrophiles such as p-(chloromethyl)benzamide,15 2-cyanobenzothiazole,16 aza-dibenzocyclooctynes,17 and perfluoro-aromatics (Fig. 1a).18–20 To date, the fastest of these catalyst-free Cys tags is a 29-residue peptide that exhibits a second-order rate constant (k) of 25.8 ± 1.8 M−1 s−1 when reacting with pentafluoro-phenyl sulfide (PS).20 Nonetheless, a rapid (k >100 M−1 s−1) system capable of site-selective protein labeling that contains a short (≤10-mer) sequence, requires no catalyst and consists of exclusively canonical residues is not yet available.



