
Affinity selection–mass spectrometry with linearizable macrocyclic peptide libraries
Abstract
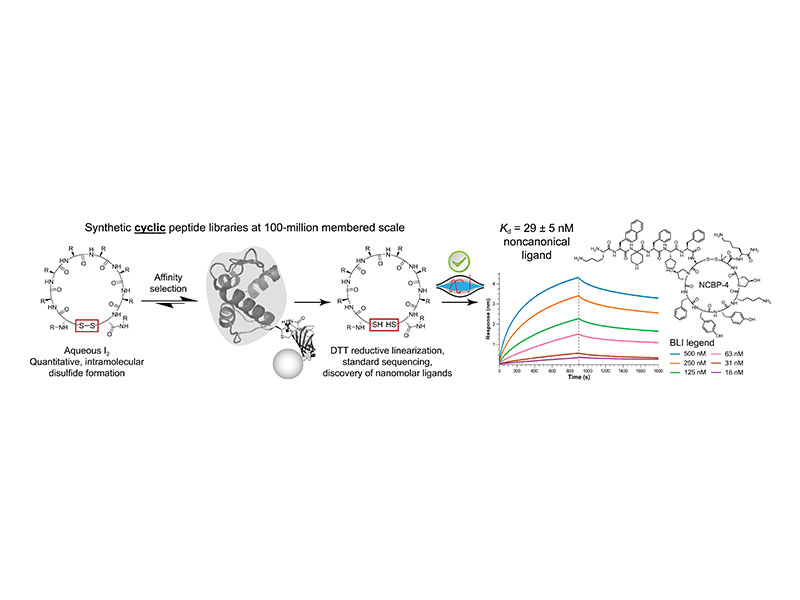
Despite their potential, the preparation of large synthetic macrocyclic libraries for ligand discovery and development has been limited. Here, we produce 100-million-membered macrocyclic libraries containing natural and nonnatural amino acids. Near-quantitative intramolecular disulfide formation is facilitated by rapid oxidation with iodine in solution. After use in affinity selection, treatment with dithiothreitol enables near-quantitative reduction, rendering linear peptide analogs for standard tandem mass spectrometry. We use these libraries to discover macrocyclic binders to cadherin-2 and anti-hemagglutinin antibody clone 12ca5. Structure-activity relationship studies of an initial cadherin-binding peptide [CBP; apparent dissociation constant (Kd) = 53 nanomolar] reveal residues responsible for driving affinity (hotspots) and mutation-tolerant residues (coldspots). Two original macrocyclic libraries are prepared in which these hotspots and coldspots are derivatized with nonnatural amino acids. Following discovery and validation, high-affinity ligands are discovered from the coldspot library, with NCBP-4 demonstrating improved affinity (Kd = 29 nanomolar). Overall, we expect that this work will improve the use of macrocyclic libraries in therapeutic peptide development.
Affinity selection–mass spectrometry with linearizable macrocyclic peptide libraries
Michael A. Lee, Joseph S. Brown, Charlotte E. Farquhar, Andrei Loas, and Bradley L. Pentelute



