
Deep Learning Enables Discovery of a Short Nuclear Targeting Peptide for Efficient Delivery of Antisense Oligomers
Deep Learning Enables Discovery of a Short Nuclear Targeting Peptide for Efficient Delivery of Antisense Oligomers
JACS Au 2021, 1, 11, 2009–2020
Publication Date:October 6, 2021
https://doi.org/10.1021/jacsau.1c00327
Eva M. López-Vidal, Carly K. Schissel, Somesh Mohapatra, Kamela Bellovoda, Chia-Ling Wu, Jenna A. Wood, Annika B. Malmberg, Andrei Loas, Rafael Gómez-Bombarelli*, and Bradley L. Pentelute
Abstract
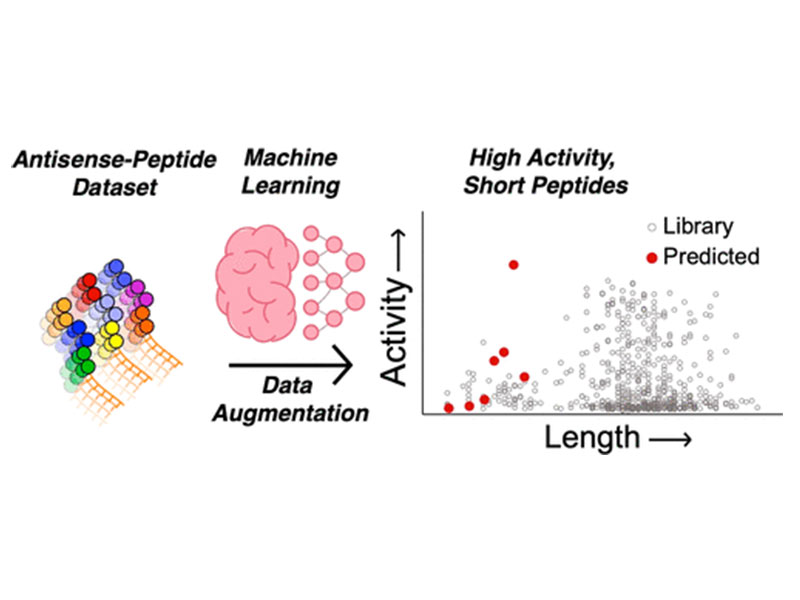
Therapeutic macromolecules such as proteins and oligonucleotides can be highly efficacious but are often limited to extracellular targets due to the cell’s impermeable membrane. Cell-penetrating peptides (CPPs) are able to deliver such macromolecules into cells, but limited structure–activity relationships and inconsistent literature reports make it difficult to design effective CPPs for a given cargo. For example, polyarginine motifs are common in CPPs, promoting cell uptake at the expense of systemic toxicity. Machine learning may be able to address this challenge by bridging gaps between experimental data in order to discern sequence–activity relationships that evade our intuition. Our earlier data set and deep learning model led to the design of miniproteins (>40 amino acids) for antisense delivery. Here, we leveraged and expanded our model with data augmentation in the short CPP sequence space of the data set to extrapolate and discover short, low-arginine-content CPPs that would be easier to synthesize and amenable to rapid conjugation to desired cargo, and with minimal in vivo toxicity. The lead predicted peptide, termed P6, is as active as a polyarginine CPP for the delivery of an antisense oligomer, while having only one arginine side chain and 18 total residues. We determined the pentalysine motif and the C-terminal cysteine of P6 to be the main drivers of activity. The antisense conjugate was able to enhance corrective splicing in an animal model to produce functional eGFP in heart tissue in vivo while remaining nontoxic up to a dose of 60 mg/kg. In addition, P6 was able to deliver an enzyme to the cytosol of cells. Our findings suggest that, given a data set of long CPPs, we can discover by extrapolation short, active sequences that deliver antisense oligomers.



